Difference between revisions of "Seongwan's Transcriptomics"
imported>Park SeongWan |
imported>Park SeongWan |
||
| Line 25: | Line 25: | ||
<h3><strong>Messenger RNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> (</span><strong>mRNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">) </span></h3> | <h3><strong>Messenger RNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> (</span><strong>mRNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">) </span></h3> | ||
| − | <p><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">is a large family of </span | + | <p><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">is a large family of </span>RNA<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> molecules that convey </span>genetic information<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> from </span>DNA<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> to the </span>ribosome<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">, where they specify the </span>amino acid<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> sequence of the </span>protein<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> products of </span>gene expression<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">. Following </span>transcription<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> of </span>primary transcript<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> mRNA (known as </span>pre-mRNA<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">) by </span>RNA polymerase<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">, processed, mature mRNA is </span>translated<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> into a polymer of amino acids.</span></p> |
<h3><span style="font-size:medium">UTR</span></h3> | <h3><span style="font-size:medium">UTR</span></h3> | ||
| Line 33: | Line 33: | ||
<h3><span style="font-size:medium">poly A</span></h3> | <h3><span style="font-size:medium">poly A</span></h3> | ||
| − | <p><strong>Polyadenylation</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> is the addition of a </span><strong>poly(A) tail</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> to a </span><a class="mw-disambig" href="http://en.wikipedia.org/wiki/Messenger" style="font-size: 14px; text-decoration: none; font-family: sans-serif; background-image: none; color: rgb(11,0,128); line-height: 22px" title="Messenger">messenger</a><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> </span> | + | <p><strong>Polyadenylation</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> is the addition of a </span><strong>poly(A) tail</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> to a </span><a class="mw-disambig" href="http://en.wikipedia.org/wiki/Messenger" style="font-size: 14px; text-decoration: none; font-family: sans-serif; background-image: none; color: rgb(11,0,128); line-height: 22px" title="Messenger">messenger</a><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> </span>RNA<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">. </span><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">The poly(A) tail is important for the nuclear export, translation, and stability of mRNA. The tail is shortened over time, and, when it is short enough, the mRNA is enzymatically degraded.</span></p> |
<h3><span style="font-size:medium">ncRNA</span></h3> | <h3><span style="font-size:medium">ncRNA</span></h3> | ||
| − | <p><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">A </span><strong>non-coding RNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> (</span><strong>ncRNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">) is a functional </span | + | <p><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">A </span><strong>non-coding RNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> (</span><strong>ncRNA</strong><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">) is a functional </span>RNA<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> molecule that is not </span>translated<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> into a </span>protein<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">. </span><span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">Non-coding RNA genes include highly abundant and functionally important RNAs such as </span>transfer RNAs<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> (tRNAs) and </span>ribosomal RNAs<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">(rRNAs), as well as RNAs such as </span>microRNAs<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">, </span>siRNAs<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px">, and </span>snRNAs.</p> |
<h1><span style="font-size:medium">measurment of RNA expression</span></h1> | <h1><span style="font-size:medium">measurment of RNA expression</span></h1> | ||
| Line 45: | Line 45: | ||
<p>RT-qPCR In this technique, reverse transcription is followed by quantitative PCR. Reverse transcription first generates a DNA template from the mRNA; this single-stranded template is called cDNA. The cDNA template is then amplified in the quantitative step, during which the fluorescence emitted by labeled hybridization probes or intercalating dyes changes as the DNA amplification process progresses.</p> | <p>RT-qPCR In this technique, reverse transcription is followed by quantitative PCR. Reverse transcription first generates a DNA template from the mRNA; this single-stranded template is called cDNA. The cDNA template is then amplified in the quantitative step, during which the fluorescence emitted by labeled hybridization probes or intercalating dyes changes as the DNA amplification process progresses.</p> | ||
| − | <p> | + | <p>RNA-seq<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> is emerging (2013) as the method of choice for measuring transcriptomes of organisms, though the older technique of </span>DNA microarrays<span style="color:rgb(37,37,37); font-family:sans-serif; font-size:14px"> is still used.</span></p> |
Revision as of 23:34, 2 June 2016
Contents
Definition
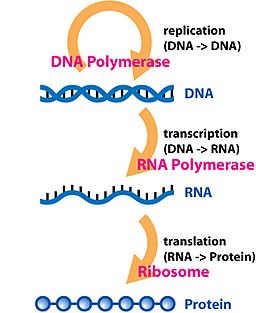
Transcriptomics is the study of the transcriptome—the complete set of RNA transcripts that are produced by the genome, under specific circumstances or in a specific cell
Relation between genomics and transcriptomics and proteomics.
Now, we can simply imagine 'central dogma'. DNA is replicated and it can be used as a template for transcription. mRNA is used as a template for translation.
The transcriptome is the product of the transcription. Therefore, genomics and transcriptomics are very related to each other. Basically, RNA is complementary to DNA.
However, RNA undergoes many different procedure, it gets its own properties.
mRNA include genetic information, codon. By the sequenced codon, ribosomes translate the mRNA into protein.
During the transcription and the translation, many regulation mechanism is existed to control the amount of expression of genome.
What are mRNAs?
Messenger RNA (mRNA)
is a large family of RNA molecules that convey genetic information from DNA to the ribosome, where they specify the amino acid sequence of the protein products of gene expression. Following transcription of primary transcript mRNA (known as pre-mRNA) by RNA polymerase, processed, mature mRNA is translated into a polymer of amino acids.
UTR
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is found on the 3' side, it is called the 3' UTR (or trailer sequence).
poly A
Polyadenylation is the addition of a poly(A) tail to a messenger RNA. The poly(A) tail is important for the nuclear export, translation, and stability of mRNA. The tail is shortened over time, and, when it is short enough, the mRNA is enzymatically degraded.
ncRNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. Non-coding RNA genes include highly abundant and functionally important RNAs such as transfer RNAs (tRNAs) and ribosomal RNAs(rRNAs), as well as RNAs such as microRNAs, siRNAs, and snRNAs.
measurment of RNA expression
Northern blotting provides size and sequence information about the mRNA molecules. A sample of RNA is separated on an agarose gel and hybridized to a radioactively labeled RNA probe that is complementary to the target sequence. The radiolabeled RNA is then detected by an autoradiograph.
RT-qPCR In this technique, reverse transcription is followed by quantitative PCR. Reverse transcription first generates a DNA template from the mRNA; this single-stranded template is called cDNA. The cDNA template is then amplified in the quantitative step, during which the fluorescence emitted by labeled hybridization probes or intercalating dyes changes as the DNA amplification process progresses.
RNA-seq is emerging (2013) as the method of choice for measuring transcriptomes of organisms, though the older technique of DNA microarrays is still used.