Difference between revisions of "About sequencing"
imported>이경준 |
imported>이경준 |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | <p> | + | <p><span style="color:#000000">In genomics, sequencing is to determine the arrangement </span><span style="color:#000000">of DNA. </span>Sequencing methods are outlined with three basic steps as shown below.</p> |
<p> </p> | <p> </p> | ||
| Line 5: | Line 5: | ||
<ol> | <ol> | ||
<li> | <li> | ||
| − | <p | + | <p>Library preparation</p> |
</li> | </li> | ||
<li> | <li> | ||
| − | <p> | + | <p>Template preparation</p> |
</li> | </li> | ||
<li> | <li> | ||
| − | <p> | + | <p>Sequencing and imaging</p> |
</li> | </li> | ||
</ol> | </ol> | ||
| Line 17: | Line 17: | ||
<p> </p> | <p> </p> | ||
| − | < | + | <h1>Library preparation</h1> |
| − | <p><span style="color:#000000">A genomic library is a collection of the total genomic DNA</span><span style="color:#000000"> from a single organism</span><span style="color:#000000">. The DNA is stored in a population of identical vectors,</span><span style="color:#000000"> each containing a different insert </span><span style="color:#000000">of DNA. In order to construct a genomic library, the organism's DNA is extracted </span><span style="color:#000000">from cells </span><span style="color:#000000">and then digested with a restriction enzyme | + | <p> </p> |
| + | |||
| + | <p><span style="color:#000000">A genomic library is a collection of the total genomic DNA</span><span style="color:#000000"> from a single organism</span><span style="color:#000000">. The DNA is stored in a population of identical vectors,</span><span style="color:#000000"> each containing a different insert </span><span style="color:#000000">of DNA. In order to construct a genomic library, the organism's DNA is extracted </span><span style="color:#000000">from cells </span><span style="color:#000000">and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. (above definition is by https://en.wikipedia.org/wiki/Genomic_library)</span></p> | ||
| + | |||
| + | <p>Fragments made by restriction enzyme undergo additional process, such as ligation of adaptor and primer, which let them be able to function in next phase.</p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
<p> </p> | <p> </p> | ||
| Line 27: | Line 35: | ||
<p> </p> | <p> </p> | ||
| − | <p>As shown as 'Good fragment', fragments are expected to have adaptor(blue colored zone in the above picture) allowing fragments to attached to | + | <p>As shown as 'Good fragment', fragments are expected to have <u>adaptor</u>(blue colored zone in the above picture) allowing fragments to attached to <strong>emPCR bead</strong> and <strong>solid phase</strong> which function for the template preparation .</p> |
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <h1>Template preparation</h1> | ||
| + | |||
| + | <p>Template preparation is to generate and amplify the template DNA, that is the main target of sequencing. there are several different ways in preparing the template DNA as much as diverse secuencing methods exist.</p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <h2>emPCR</h2> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p><img alt="" src="/ckfinder/userfiles/images/emPCR%2C%20Solid-phase%20amplification%20(2).png" style="height:183px; width:731px" /></p> | ||
| + | |||
| + | <p><strong><em> <Summary of the Em PCR></em></strong></p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p>There are primer, template DNA, dNTPs, DNA polymerase mixed with emulsion oil and water. </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p><img alt="" src="/ckfinder/userfiles/images/emulsion%20(2).jpg" style="height:139px; width:404px" /></p> | ||
| + | |||
| + | <p>After shaking to make emulsion, the PCR mix enters Into the emulsion(the oil dorp) and denaturation begins to seperate double strands.</p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p><img alt="" src="/ckfinder/userfiles/images/2(2).jpg" style="height:389px; width:550px" /></p> | ||
| + | |||
| + | <p>Let's call the fragment attached to the bead with adaptod(colored blue) 'reverse strand' for conveneince.</p> | ||
| + | |||
| + | <p>When the reverse strand annealed to the bead, polymerase amplifies the forward strand from the bead so that it produce one 'forward strand'.</p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p><img alt="" src="/ckfinder/userfiles/images/3(2).jpg" style="height:376px; width:550px" /></p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p>Then the 'reverse strand', which was annealed to the bead, denatured and annealed again to the other spot.</p> | ||
| + | |||
| + | <p>And the primer also anneals to the forward strand, which was produced before.</p> | ||
| + | |||
| + | <p><img alt="" src="/ckfinder/userfiles/images/4(2).jpg" style="height:405px; width:550px" /></p> | ||
| + | |||
| + | <p>Repeat these processes so far, for 30-60 cycles and generate and amplify the template DNA.</p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <h3>Quetions</h3> | ||
| + | |||
| + | <p>Why is the oil required?</p> | ||
| + | |||
| + | <p>Why is the sphere-shape bead required?</p> | ||
<p> </p> | <p> </p> | ||
| Line 33: | Line 115: | ||
<p> </p> | <p> </p> | ||
| − | <p> | + | <h2>Solid-phase</h2> |
| + | |||
| + | <p><img alt="" src="/ckfinder/userfiles/images/emPCR%2C%20Solid-phase%20amplification%20(3).png" style="height:285px; width:478px" /></p> | ||
| + | |||
| + | <p><summary of the solid-phase amplification></p> | ||
| + | |||
| + | <p>Firstly attach the primers(adaptor) on the slide and input the prepared sample that has adaptor to attach on the slide as well. </p> | ||
| + | |||
| + | <p>When the sample attach to one primer on the slide, it bent over and allow the another end of sample fragment to attach to other primer on the slide. Then polymerizing starts.</p> | ||
| + | |||
| + | <p>This cycle is repeated and make clusters. </p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p>I'd like you to refer this video about Illumina sequencing, which includes solid phase amplification phase.</p> | ||
| + | |||
| + | <p>Illumina sequencing https://www.youtube.com/watch?v=womKfikWlxM</p> | ||
<p> </p> | <p> </p> | ||
| Line 39: | Line 137: | ||
<p> </p> | <p> </p> | ||
| − | <p>Sequencing and imaging</p> | + | <h1>Sequencing and imaging</h1> |
| + | |||
| + | <p><img alt="Principles for sequencing and imaging. (a) Illumina sequencing of three template molecules. All four nucleotides, carrying terminating moieties and unique fluorescent labels, and DNA polymerase are added, and one complementary nucleotide becomes incorporated at each template molecule (1). After washing, fluorescence is registered at four wavelengths (2). Fluorescent dyes and terminating groups are cleaved off. A new set of nucleotides is added (3), and imaged (4). Sequence reads of equal length are obtained (5). (b) 454 sequencing of three template molecules. One type of natural non-terminating deoxynucleotides and DNA polymerase are added and a pyrophosphate molecule is released at each nucleotide incorporation (1). Pyrophosphate is converted into light using sulfurylase and luciferase, and the light intensity is measured in each well (2). Free deoxynucleotides are destroyed with apyrase before adding the next type of deoxynucleotide (3) and imaging (4). Light signals are converted to flowgrams with higher signal intensity bars in homopolymer regions (5). Sequence reads that may differ in length are obtained (6). (c) SOLiD sequencing of one template molecule. A sequencing primer, DNA ligase and 1,024 unique probes, which are fluorescently labeled according to their first two bases, are added, and the complementary probe is ligated to the template (1). After washing, fluorescence is registered at four wavelengths. The three universal bases and the fluorophor are cleaved off (2). Addition of a new probe set is repeated for the desired number of cycles (3,4). The newly built strand is melted off. A new sequencing primer is added, which anneals one base off from the first primer and therefore interrogates different positions (5). Sequencing is repeated for the desired number of cycles (6). Additional primers are added, until each base is sequenced twice. The colors from all sequencing rounds are merged and can be converted to nucleotides (7)." src="https://openi.nlm.nih.gov/imgs/512/340/3267688/PMC3267688_2041-2223-2-23-3.png" /></p> | ||
| + | |||
| + | <p><strong> </strong>Principles for sequencing and imaging.</p> | ||
| + | |||
| + | <p>(a) Illumina sequencing of three template molecules. All four nucleotides, carrying terminating moieties and unique fluorescent labels, and DNA polymerase are added, and one complementary nucleotide becomes incorporated at each template molecule (1). After washing, fluorescence is registered at four wavelengths (2). Fluorescent dyes and terminating groups are cleaved off. A new set of nucleotides is added (3), and imaged (4). Sequence reads of equal length are obtained (5).</p> | ||
| + | |||
| + | <p>(b) 454 sequencing of three template molecules. One type of natural non-terminating deoxynucleotides and DNA polymerase are added and a pyrophosphate molecule is released at each nucleotide incorporation (1). Pyrophosphate is converted into light using sulfurylase and luciferase, and the light intensity is measured in each well (2). Free deoxynucleotides are destroyed with apyrase before adding the next type of deoxynucleotide (3) and imaging (4). Light signals are converted to flowgrams with higher signal intensity bars in homopolymer regions (5). Sequence reads that may differ in length are obtained (6).</p> | ||
| + | |||
| + | <p>(c) SOLiD sequencing of one template molecule. A sequencing primer, DNA ligase and 1,024 unique probes, which are fluorescently labeled according to their first two bases, are added, and the complementary probe is ligated to the template (1). After washing, fluorescence is registered at four wavelengths. The three universal bases and the fluorophor are cleaved off (2). Addition of a new probe set is repeated for the desired number of cycles (3,4). The newly built strand is melted off. A new sequencing primer is added, which anneals one base off from the first primer and therefore interrogates different positions (5). Sequencing is repeated for the desired number of cycles (6). Additional primers are added, until each base is sequenced twice. The colors from all sequencing rounds are merged and can be converted to nucleotides (7).</p> | ||
| + | |||
| + | <h1>Reference</h1> | ||
| + | |||
| + | <p>Figures</p> | ||
| + | |||
| + | <p>https://www.nature.com/articles/nrg2626</p> | ||
| + | |||
| + | <p>https://openi.nlm.nih.gov/detailedresult.php?img=PMC3267688_2041-2223-2-23-3&req=4 </p> | ||
Latest revision as of 10:38, 15 December 2017
In genomics, sequencing is to determine the arrangement of DNA. Sequencing methods are outlined with three basic steps as shown below.
-
Library preparation
-
Template preparation
-
Sequencing and imaging
Contents
Library preparation
A genomic library is a collection of the total genomic DNA from a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. (above definition is by https://en.wikipedia.org/wiki/Genomic_library)
Fragments made by restriction enzyme undergo additional process, such as ligation of adaptor and primer, which let them be able to function in next phase.
As shown as 'Good fragment', fragments are expected to have adaptor(blue colored zone in the above picture) allowing fragments to attached to emPCR bead and solid phase which function for the template preparation .
Template preparation
Template preparation is to generate and amplify the template DNA, that is the main target of sequencing. there are several different ways in preparing the template DNA as much as diverse secuencing methods exist.
emPCR
<Summary of the Em PCR>
There are primer, template DNA, dNTPs, DNA polymerase mixed with emulsion oil and water.
After shaking to make emulsion, the PCR mix enters Into the emulsion(the oil dorp) and denaturation begins to seperate double strands.
Let's call the fragment attached to the bead with adaptod(colored blue) 'reverse strand' for conveneince.
When the reverse strand annealed to the bead, polymerase amplifies the forward strand from the bead so that it produce one 'forward strand'.
Then the 'reverse strand', which was annealed to the bead, denatured and annealed again to the other spot.
And the primer also anneals to the forward strand, which was produced before.
Repeat these processes so far, for 30-60 cycles and generate and amplify the template DNA.
Quetions
Why is the oil required?
Why is the sphere-shape bead required?
Solid-phase
<summary of the solid-phase amplification>
Firstly attach the primers(adaptor) on the slide and input the prepared sample that has adaptor to attach on the slide as well.
When the sample attach to one primer on the slide, it bent over and allow the another end of sample fragment to attach to other primer on the slide. Then polymerizing starts.
This cycle is repeated and make clusters.
I'd like you to refer this video about Illumina sequencing, which includes solid phase amplification phase.
Illumina sequencing https://www.youtube.com/watch?v=womKfikWlxM
Sequencing and imaging
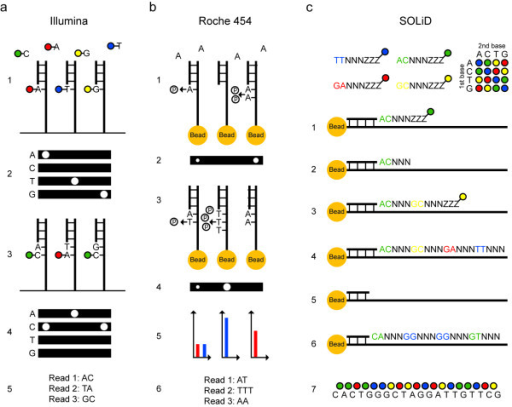
Principles for sequencing and imaging.
(a) Illumina sequencing of three template molecules. All four nucleotides, carrying terminating moieties and unique fluorescent labels, and DNA polymerase are added, and one complementary nucleotide becomes incorporated at each template molecule (1). After washing, fluorescence is registered at four wavelengths (2). Fluorescent dyes and terminating groups are cleaved off. A new set of nucleotides is added (3), and imaged (4). Sequence reads of equal length are obtained (5).
(b) 454 sequencing of three template molecules. One type of natural non-terminating deoxynucleotides and DNA polymerase are added and a pyrophosphate molecule is released at each nucleotide incorporation (1). Pyrophosphate is converted into light using sulfurylase and luciferase, and the light intensity is measured in each well (2). Free deoxynucleotides are destroyed with apyrase before adding the next type of deoxynucleotide (3) and imaging (4). Light signals are converted to flowgrams with higher signal intensity bars in homopolymer regions (5). Sequence reads that may differ in length are obtained (6).
(c) SOLiD sequencing of one template molecule. A sequencing primer, DNA ligase and 1,024 unique probes, which are fluorescently labeled according to their first two bases, are added, and the complementary probe is ligated to the template (1). After washing, fluorescence is registered at four wavelengths. The three universal bases and the fluorophor are cleaved off (2). Addition of a new probe set is repeated for the desired number of cycles (3,4). The newly built strand is melted off. A new sequencing primer is added, which anneals one base off from the first primer and therefore interrogates different positions (5). Sequencing is repeated for the desired number of cycles (6). Additional primers are added, until each base is sequenced twice. The colors from all sequencing rounds are merged and can be converted to nucleotides (7).
Reference
Figures
https://www.nature.com/articles/nrg2626
https://openi.nlm.nih.gov/detailedresult.php?img=PMC3267688_2041-2223-2-23-3&req=4