Difference between revisions of "Week 2 YY"
imported>Youyoung Kim (Created page with "<p><span style="color:#FFFFFF"><span style="font-size:16px"><span style="font-family:comic sans ms,cursive"><span style="background-color:#000080">What is S<strong>equencing</str...") |
imported>Youyoung Kim |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<p><span style="font-family:comic sans ms,cursive"><span style="font-size:14px">DNA sequencing is the process of determining the nucleotide order of a given fragment.</span></span></p> | <p><span style="font-family:comic sans ms,cursive"><span style="font-size:14px">DNA sequencing is the process of determining the nucleotide order of a given fragment.</span></span></p> | ||
| − | <p> | + | <p> </p> |
| − | <p>Sanger, NGS, | + | <p><span style="font-family:comic sans ms,cursive"><span style="font-size:14px">1. Sanger sequencing</span></span></p> |
| + | |||
| + | <p><img alt="sanger sequencing에 대한 이미지 검색결과" src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/3d/Radioactive_Fluorescent_Seq.jpg/220px-Radioactive_Fluorescent_Seq.jpg" /></p> | ||
| + | |||
| + | <p><span style="font-size:14px"><span style="font-family:comic sans ms,cursive">Sanger sequencing is a method of DNA sequencing based on the selective incorporation of chain-terminating dideoxynucleotides(ddNTPs) by DNA polymerase during in vitro DNA replication. This method was developed by Fredric Sanger and colleagues in 1977. </span></span></p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p><span style="font-size:14px"><span style="font-family:comic sans ms,cursive">2. Next-generation sequencing (NGS)</span></span></p> | ||
| + | |||
| + | <p><img alt="next generation sequencing에 대한 이미지 검색결과" src="http://image.slidesharecdn.com/introductiontonextgenerationsequencingv2-110927082709-phpapp02/95/introduction-to-next-generation-sequencing-29-728.jpg?cb=1317179320" style="height:400px; width:350px" /></p> | ||
| + | |||
| + | <p><span style="font-size:14px"><span style="font-family:comic sans ms,cursive">Next-generation sequencing is the catch-all term used to describe a number of different modern sequencing technologies including Illumina sequencing, Roche 454 sequencing, Ion torrent: Proton/ PGM sequencing, and SOLiD sequencing. These recent technologies allow us to sequence DNA and RNA much more quickly and cheaply than the previously used Sanger sequencing, and as such have revolutionised the study of genomics and molecular biology.</span></span></p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p><span style="font-size:14px"><span style="font-family:comic sans ms,cursive">In addition, there are other types of sequencing such as methyl-seq or ATAC-seq.</span></span></p> | ||
| + | |||
| + | <p> </p> | ||
Latest revision as of 23:59, 30 November 2016
What is Sequencing?
DNA sequencing is the process of determining the nucleotide order of a given fragment.
1. Sanger sequencing
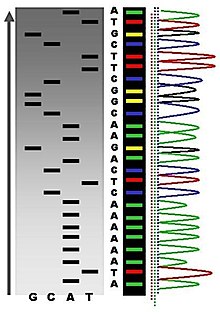
Sanger sequencing is a method of DNA sequencing based on the selective incorporation of chain-terminating dideoxynucleotides(ddNTPs) by DNA polymerase during in vitro DNA replication. This method was developed by Fredric Sanger and colleagues in 1977.
2. Next-generation sequencing (NGS)

Next-generation sequencing is the catch-all term used to describe a number of different modern sequencing technologies including Illumina sequencing, Roche 454 sequencing, Ion torrent: Proton/ PGM sequencing, and SOLiD sequencing. These recent technologies allow us to sequence DNA and RNA much more quickly and cheaply than the previously used Sanger sequencing, and as such have revolutionised the study of genomics and molecular biology.
In addition, there are other types of sequencing such as methyl-seq or ATAC-seq.