Difference between pages "Chapter !9 - Microarrays and Transcriptomics Code : KSI0018" and "Essay !1 - Genomics & Omics - Code : KSI0001"
imported>김상인 |
imported>김상인 |
||
| Line 1: | Line 1: | ||
| − | <p | + | <p> </p> |
<p> </p> | <p> </p> | ||
| − | < | + | <h2>You Will Learn</h2> |
| − | < | + | <ul> |
| + | <li>The principles of genetics, genes and traits</li> | ||
| + | <li>The applications and implications of genome sequencing</li> | ||
| + | <li>How personal genomics might impact healthcare</li> | ||
| + | <li>Tools used to diagnose and treat diseases</li> | ||
| + | <li>Methods for determining the heritability of traits and diseases</li> | ||
| + | </ul> | ||
| − | <p> | + | <p><strong>Genomics</strong> refers to the study of the <a href="https://en.wikipedia.org/wiki/Genome" title="Genome">genome</a><sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-gov2010a-1">[1]</a></sup> in contrast to <a href="https://en.wikipedia.org/wiki/Genetics" title="Genetics">genetics</a> which refers to the study of genes and their roles in inheritance.<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-gov2010a-1">[1]</a></sup> Genomics can be considered a discipline in genetics. It applies <a href="https://en.wikipedia.org/wiki/Recombinant_DNA" title="Recombinant DNA">recombinant DNA</a>, <a href="https://en.wikipedia.org/wiki/DNA_sequencing" title="DNA sequencing">DNA sequencing</a> methods, and <a href="https://en.wikipedia.org/wiki/Bioinformatics" title="Bioinformatics">bioinformatics</a> to sequence, assemble, and analyze the function and structure of genomes (the <em>complete</em> set of DNA within a single cell of an organism).<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-gov2010-2">[2]</a></sup><sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-Klug_2012-3">[3]</a></sup> Advances in genomics have triggered a revolution in discovery-based research to understand even the most complex biological systems such as the brain.<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-Kadakkuzha_2013-4">[4]</a></sup> The field includes efforts to determine the entire <a href="https://en.wikipedia.org/wiki/DNA_sequence" title="DNA sequence">DNA sequence</a> of organisms and fine-scale <a href="https://en.wikipedia.org/wiki/Genetic_mapping" title="Genetic mapping">genetic mapping</a>. The field also includes studies of intragenomic phenomena such as <a href="https://en.wikipedia.org/wiki/Heterosis" title="Heterosis">heterosis</a>, <a href="https://en.wikipedia.org/wiki/Epistasis" title="Epistasis">epistasis</a>, <a href="https://en.wikipedia.org/wiki/Pleiotropy" title="Pleiotropy">pleiotropy</a> and other interactions between <a href="https://en.wikipedia.org/wiki/Locus_(genetics)" title="Locus (genetics)">loci</a> and <a href="https://en.wikipedia.org/wiki/Allele" title="Allele">alleles</a> within the genome.<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-Pevsner_2009-5">[5]</a></sup> In contrast, the investigation of the roles and functions of single genes is a primary focus of <a href="https://en.wikipedia.org/wiki/Molecular_biology" title="Molecular biology">molecular biology</a> or <a href="https://en.wikipedia.org/wiki/Genetics" title="Genetics">genetics</a> and is a common topic of modern medical and biological research. Research carried out into single genes does not generally fall into the definition of genomics unless the aim of this genetic, pathway, and functional information analysis is to elucidate its effect on, place in, and response to the entire genomes networks.<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-Culver_2002-6">[6]</a></sup><sup>[<em><a href="https://en.wikipedia.org/wiki/Wikipedia:Verifiability" title="Wikipedia:Verifiability">not specific enough to verify</a></em>]</sup></p> |
| + | |||
| + | <p> </p> | ||
| − | <p> | + | <p> </p> |
| − | < | + | <h3>The "omics" revolution[<a href="https://en.wikipedia.org/w/index.php?title=Genomics&action=edit&section=6" title="Edit section: The "omics" revolution">edit</a>]</h3> |
| − | <p> </p> | + | <p>Main articles: <a href="https://en.wikipedia.org/wiki/Omics" title="Omics">Omics</a> and <a href="https://en.wikipedia.org/wiki/Human_proteome_project" title="Human proteome project">Human proteome project</a></p> |
| − | <p> | + | <p>The English-language <a href="https://en.wikipedia.org/wiki/Neologism" title="Neologism">neologism</a> <strong>omics</strong> informally refers to a field of study in <a href="https://en.wikipedia.org/wiki/Biology" title="Biology">biology</a> ending in <em>-omics</em>, such as genomics, <a href="https://en.wikipedia.org/wiki/Proteomics" title="Proteomics">proteomics</a> or <a href="https://en.wikipedia.org/wiki/Metabolomics" title="Metabolomics">metabolomics</a>. The related suffix <strong>-ome</strong> is used to address the objects of study of such fields, such as the <a href="https://en.wikipedia.org/wiki/Genome" title="Genome">genome</a>, <a href="https://en.wikipedia.org/wiki/Proteome" title="Proteome">proteome</a> or <a href="https://en.wikipedia.org/wiki/Metabolome" title="Metabolome">metabolome</a> respectively. The suffix <em>-ome</em> as used in molecular biology refers to a <em>totality</em> of some sort; similarly <strong>omics</strong> has come to refer generally to the study of large, comprehensive biological data sets. While the growth in the use of the term has led some scientists (<a href="https://en.wikipedia.org/wiki/Jonathan_Eisen" title="Jonathan Eisen">Jonathan Eisen</a>, among others<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-Eisen_2012-41">[41]</a></sup>) to claim that it has been oversold,<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-wsj_2012-42">[42]</a></sup> it reflects the change in orientation towards the quantitative analysis of complete or near-complete assortment of all the constituents of a system.<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-scudellari2011-43">[43]</a></sup> In the study of <a href="https://en.wikipedia.org/wiki/Symbiosis" title="Symbiosis">symbioses</a>, for example, researchers which were once limited to the study of a single gene product can now simultaneously compare the total complement of several types of biological molecules.<sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-Chaston_2012-44">[44]</a></sup><sup><a href="https://en.wikipedia.org/wiki/Genomics#cite_note-McCutcheon_2011-45">[45]</a></sup></p> |
<p> </p> | <p> </p> | ||
| − | <p>& | + | <p>The English-language <a href="https://en.wikipedia.org/wiki/Neologism" title="Neologism">neologism</a> <strong>omics</strong> informally refers to a field of study in <a href="https://en.wikipedia.org/wiki/Biology" title="Biology">biology</a> ending in <em>-omics</em>, such as <a href="https://en.wikipedia.org/wiki/Genomics" title="Genomics">genomics</a>, <a href="https://en.wikipedia.org/wiki/Proteomics" title="Proteomics">proteomics</a> or <a href="https://en.wikipedia.org/wiki/Metabolomics" title="Metabolomics">metabolomics</a>. The related suffix <strong>-ome</strong> is used to address the objects of study of such fields, such as the <a href="https://en.wikipedia.org/wiki/Genome" title="Genome">genome</a>, <a href="https://en.wikipedia.org/wiki/Proteome" title="Proteome">proteome</a> or <a href="https://en.wikipedia.org/wiki/Metabolome" title="Metabolome">metabolome</a> respectively. Omics aims at the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms.</p> |
| − | <p> | + | <p><a href="https://en.wikipedia.org/wiki/Functional_genomics" title="Functional genomics">Functional genomics</a> aims at identifying the functions of as many genes as possible of a given organism. It combines different -omics techniques such as transcriptomics and proteomics with saturated mutant collections.<sup><a href="https://en.wikipedia.org/wiki/Omics#cite_note-1">[1]</a></sup></p> |
| − | <p> | + | <p>The suffix <em>-ome</em> as used in molecular biology refers to a <em>totality</em> of some sort; it is an example of a "neo-suffix" formed by abstraction from various Greek terms in -ωμα, a sequence that does not form an identifiable suffix in Greek.</p> |
| − | <p> | + | <p> </p> |
<p> </p> | <p> </p> | ||
| − | < | + | <h2>Current usage[<a href="https://en.wikipedia.org/w/index.php?title=Omics&action=edit&section=12" title="Edit section: Current usage">edit</a>]</h2> |
| − | <p> | + | <p>Main article: <a href="https://en.wikipedia.org/wiki/List_of_omics_topics_in_biology" title="List of omics topics in biology">List of omics topics in biology</a></p> |
| + | |||
| + | <p>Many “omes” beyond the original “<a href="https://en.wikipedia.org/wiki/Genome" title="Genome">genome</a>” have become useful and have been widely adopted by research scientists. “<a href="https://en.wikipedia.org/wiki/Proteomics" title="Proteomics">Proteomics</a>” has become well-established as a term for studying <a href="https://en.wikipedia.org/wiki/Proteins" title="Proteins">proteins</a> at a large scale. "Omes" can provide an easy shorthand to encapsulate a field; for example, an <a href="https://en.wikipedia.org/wiki/Interactomics" title="Interactomics">interactomics</a> study is clearly recognisable as relating to large-scale analyses of gene-gene, protein-protein, or protein-ligand interactions. Researchers are rapidly taking up omes and omics, as shown by the explosion of the use of these terms in <a href="https://en.wikipedia.org/wiki/PubMed" title="PubMed">PubMed</a> since the mid '90s.<sup><a href="https://en.wikipedia.org/wiki/Omics#cite_note-10">[10]</a></sup></p> | ||
<p> </p> | <p> </p> | ||
| − | <p> | + | <p><img alt="" src="/ckfinder/userfiles/images/genomics%20omics.PNG" style="height:217px; width:1132px" /></p> |
| − | <p> | + | <p> </p> |
| − | <p> | + | <p>Genomics is the new science that deals with the discovery and noting of all the sequences in the entire genome of a particular organism. The genome can be defined as the complete set of genes inside a cell. Genomics, is, therefore, the study of the genetic make-up of organisms.</p> |
| − | <p>- | + | <p>Determining the genomic sequence, however, is only the beginning of genomics. Once this is done, the genomic sequence is used to study the function of the numerous genes (functional genomics), to compare the genes in one organism with those of another (comparative genomics), or to generate the 3-D structure of one or more proteins from each protein family, thus offering clues to their function (structural genomics).</p> |
| − | <p> | + | <p>In crop agriculture, the main purpose of the application of genomics is to gain a better understanding of the whole genome of plants. Agronomically important genes may be identified and targeted to produce more nutritious and safe food while at the same time preserving the environment.</p> |
| − | <p>& | + | <p>Genomics is an entry point for looking at the other ‘omics’ sciences. The information in the genes of an organism, its genotype, is largely responsible for the final physical makeup of the organism, referred to as the “phenotype”. However, the environment also has some influence on the phenotype.</p> |
| − | <p>& | + | <p>DNA in the genome is only one aspect of the complex mechanism that keeps an organism running – so decoding the DNA is one step towards understanding the process. However, by itself, it does not specify everything that happens within the organism.</p> |
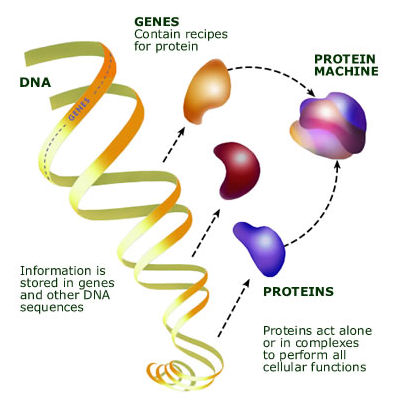
| − | <p>& | + | <p>The basic flow of genetic information in a cell is as follows. The DNA is transcribed or copied into a form known as “RNA”. The complete set of RNA (also known as its transcriptome) is subject to some editing (cutting and pasting) to become messenger-RNA, which carries information to the ribosome, the protein factory of the cell, which then translates the message into protein.</p> |
| − | <p>& | + | <table border="0" cellpadding="0" cellspacing="0" style="width:405px"> |
| + | <tbody> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p>Figure 1. Genes, proteins, and molecular machines</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p><img src="http://www.isaaa.org/siteimages/pocketkimages/clip_image002_0014.jpg" style="height:416px; width:405px" /></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p>Source: U.S. Department of Energy Genomes to Life Program, <a href="http://doegenomestolife.org/" target="_blank">http://doegenomestolife.org</a></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| − | < | + | <h3>The International Rice Genome Sequencing Project</h3> |
| − | <p> | + | <table align="left" border="0" cellpadding="5" cellspacing="0"> |
| + | <tbody> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p><img src="http://www.isaaa.org/siteimages/pocketkimages/clip_image003_0012.jpg" style="height:82px; width:125px" /></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| − | <p>& | + | <p>This ongoing genomic research in rice is a collaborative effort of several public and private laboratories worldwide. This project aims to completely sequence the entire rice genome (12 rice chromosomes) and subsequently apply the knowledge to improve rice production.<br /> |
| + | In 2002, the draft genome sequences of two agriculturally important subspecies of rice, indica and japonica, were published. Once completed, the rice genome sequence will serve as a model system for other cereal grasses and will assist in identifying important genes in maize, wheat, oats, sorghum, and millet.<br /> | ||
| + | For more, visit <a href="http://rgp.dna.affrc.go.jp/IRGSP" target="_blank">http://rgp.dna.affrc.go.jp/IRGSP</a></p> | ||
| − | < | + | <h2>Proteomics</h2> |
| − | <p> | + | <p>Proteins are responsible for an endless number of tasks within the cell. The complete set of proteins in a cell can be referred to as its proteome and the study of protein structure and function and what every protein in the cell is doing is known as proteomics. The proteome is highly dynamic and it changes from time to time in response to different environmental stimuli. The goal of proteomics is to understand how the structure and function of proteins allow them to do what they do, what they interact with, and how they contribute to life processes.</p> |
| − | <p> | + | <p>An application of proteomics is known as protein “expression profiling” where proteins are identified at a certain time in an organism as a result of the expression to a stimulus. Proteomics can also be used to develop a protein-network map where interaction among proteins can be determined for a particular living system.</p> |
| − | <p> | + | <p>Proteomics can also be applied to map protein modification to determine the difference between a wild type and a genetically modified organism. It is also used to study protein-protein interactions involved in plant defense reactions.</p> |
| − | <p> | + | <table align="right" border="0" cellpadding="5" cellspacing="0"> |
| + | <tbody> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p><img src="http://www.isaaa.org/siteimages/pocketkimages/clip_image004_0011.jpg" style="height:82px; width:125px" /></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| − | <p> | + | <p>For example, proteomics research at Iowa State University, USA includes:</p> |
| − | < | + | <ul> |
| + | <li>an examination of changes of protein in the corn proteome during low temperatures which is a major problem for young corn seedlings;</li> | ||
| + | <li>analysis of the differences that occur in the genome expression in developing soybean stressed by high temperatures; and</li> | ||
| + | <li>identifying the proteins expressed in response to diseases like soybean cyst nematode.</li> | ||
| + | </ul> | ||
| − | < | + | <h2>Metabolomics</h2> |
| − | <p>& | + | <p>Metabolomics is one of the newest ‘omics’ sciences. The metabolome refers to the complete set of low molecular weight compounds in a sample. These compounds are the substrates and by-products of enzymatic reactions and have a direct effect on the phenotype of the cell. Thus, metabolomics aims at determining a sample’s profile of these compounds at a specified time under specific environmental conditions.</p> |
| − | <p> | + | <p>Genomics and proteomics have provided extensive information regarding the genotype but convey limited information about phenotype. Low molecular weight compounds are the closest link to phenotype.</p> |
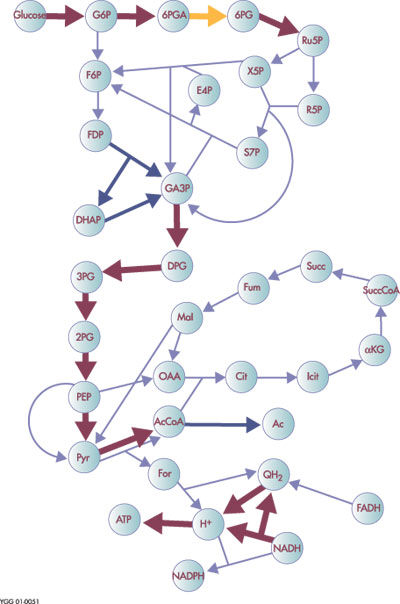
| − | <p> | + | <p>Metabolomics can be used to determine differences between the levels of thousands of molecules between a healthy and diseased plant. The technology can also be used to determine the nutritional difference between traditional and genetically modified crops, and in identifying plant defense metabolites.</p> |
| − | <p>2. | + | <table border="0" cellpadding="0" cellspacing="0" style="width:405px"> |
| + | <tbody> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p>Figure 2. Example of a metabolic network model for <em>E. coli</em></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p><img src="http://www.isaaa.org/siteimages/pocketkimages/clip_image005_0003.jpg" style="height:604px; width:400px" /></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p>Source: U.S. Department of Energy Genomes to Life Program, <a href="http://doegenomestolife.org/" target="_blank">http://doegenomestolife.org</a></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
<p> </p> | <p> </p> | ||
| − | <p> | + | <table border="0" cellpadding="0" cellspacing="0" style="width:405px"> |
| + | <tbody> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p>Figure 2. Example of a metabolic network model for <em>E. coli</em></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p><img src="http://www.isaaa.org/siteimages/pocketkimages/clip_image006_0000.jpg" style="height:240px; width:400px" /></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p>Source: <a href="http://biotech.nature.com/" target="_blank">http://biotech.nature.com</a></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| − | <p>& | + | <p> </p> |
| + | |||
| + | <p>Genomics provides an overview of the complete set of genetic instructions provided by the DNA, while transcriptomics looks into gene expression patterns. Proteomics studies dynamic protein products and their interactions, while metabolomics is also an intermediate step in understanding organism’s entire metabolism.</p> | ||
<p> </p> | <p> </p> | ||
| − | < | + | <h2>Glossary</h2> |
| − | <p | + | <p><strong>Chromosome</strong>: a grouping of coiled strands of DNA, containing many genes.</p> |
| − | <p> | + | <p><strong>DNA (deoxyribonucleic acid)</strong>: a molecule found in cells of organisms that encodes genetic information.</p> |
| − | <p> | + | <p><strong>Gene</strong>: a biological unit that codes for distinct traits or characteristics.</p> |
| − | <p> | + | <p><strong>Genome</strong>: the complete set of genes in a cell.</p> |
| − | <p | + | <p><strong>Genotype</strong>: the genetic constitution of an organism.</p> |
| − | <p> | + | <p><strong>Metabolome</strong>: complete set of low molecular weight compounds in a cell at a given time.</p> |
| − | <p> | + | <p><strong>Phenotype</strong>: the physical appearance/observable characteristics of an organism.</p> |
| − | <p> | + | <p><strong>Proteome</strong>: complete set of proteins in a cell at a given time.</p> |
| − | <p>< | + | <p><strong>RNA (ribonucleic acid)</strong>: a molecule, derived from DNA by transcription, that either carries information (messenger RNA), provides sub-cellular structure (ribosomal RNA), transports amino acids (transfer RNA), or facilitates the biochemical modification of itself or other RNA molecules.</p> |
| − | < | + | <h2>References</h2> |
| − | < | + | <ol> |
| + | <li>Genomics and Its impact on Medicine and Society. A 2001 primer. <a href="http://www.ornl.gov/TechResources/Human_Genome/publicat/primer2001/1.html" target="_blank">http://www.ornl.gov/TechResources/Human_Genome/publicat/primer2001/1.html</a></li> | ||
| + | <li>Primer on Molecular Genetics. <a href="http://www.ornl.gov/TechResources/Human_Genome/publicat/primer/primer.pdf" target="_blank">http://www.ornl.gov/TechResources/Human_Genome/publicat/primer/primer.pdf</a></li> | ||
| + | <li>Plant Sciences Institute Update. Iowa State University. October 2001. Volume 2 No.1.</li> | ||
| + | <li>Genome Projects. <a href="http://www.tigr.org/tdb" target="_blank">http://www.tigr.org/tdb</a></li> | ||
| + | <li>Meet the ‘omics’ 2003 Agbiotech Infosource. Saskatchewan Agricultural Biotechnology Information Centre, A service of Ag-West Biotech Inc.</li> | ||
| + | </ol> | ||
| − | <p> | + | <p>Reference</p> |
| − | <p> </p> | + | <p>1. https://en.wikipedia.org/wiki/Genomics</p> |
| − | <p> | + | <p>2. https://en.wikipedia.org/wiki/Omics</p> |
| − | <p> | + | <p>3. http://www.isaaa.org/resources/publications/pocketk/15/default.asp</p> |
| − | <p>- | + | <p>4. http://geneticscertificate.stanford.edu/courses/genomics-and-the-other-omics</p> |
<p> </p> | <p> </p> | ||
| − | <p> | + | <p> </p> |
<p> </p> | <p> </p> | ||
Revision as of 23:04, 30 November 2016
Contents
You Will Learn
- The principles of genetics, genes and traits
- The applications and implications of genome sequencing
- How personal genomics might impact healthcare
- Tools used to diagnose and treat diseases
- Methods for determining the heritability of traits and diseases
Genomics refers to the study of the genome[1] in contrast to genetics which refers to the study of genes and their roles in inheritance.[1] Genomics can be considered a discipline in genetics. It applies recombinant DNA, DNA sequencing methods, and bioinformatics to sequence, assemble, and analyze the function and structure of genomes (the complete set of DNA within a single cell of an organism).[2][3] Advances in genomics have triggered a revolution in discovery-based research to understand even the most complex biological systems such as the brain.[4] The field includes efforts to determine the entire DNA sequence of organisms and fine-scale genetic mapping. The field also includes studies of intragenomic phenomena such as heterosis, epistasis, pleiotropy and other interactions between loci and alleles within the genome.[5] In contrast, the investigation of the roles and functions of single genes is a primary focus of molecular biology or genetics and is a common topic of modern medical and biological research. Research carried out into single genes does not generally fall into the definition of genomics unless the aim of this genetic, pathway, and functional information analysis is to elucidate its effect on, place in, and response to the entire genomes networks.[6][not specific enough to verify]
The "omics" revolution[edit]
Main articles: Omics and Human proteome project
The English-language neologism omics informally refers to a field of study in biology ending in -omics, such as genomics, proteomics or metabolomics. The related suffix -ome is used to address the objects of study of such fields, such as the genome, proteome or metabolome respectively. The suffix -ome as used in molecular biology refers to a totality of some sort; similarly omics has come to refer generally to the study of large, comprehensive biological data sets. While the growth in the use of the term has led some scientists (Jonathan Eisen, among others[41]) to claim that it has been oversold,[42] it reflects the change in orientation towards the quantitative analysis of complete or near-complete assortment of all the constituents of a system.[43] In the study of symbioses, for example, researchers which were once limited to the study of a single gene product can now simultaneously compare the total complement of several types of biological molecules.[44][45]
The English-language neologism omics informally refers to a field of study in biology ending in -omics, such as genomics, proteomics or metabolomics. The related suffix -ome is used to address the objects of study of such fields, such as the genome, proteome or metabolome respectively. Omics aims at the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms.
Functional genomics aims at identifying the functions of as many genes as possible of a given organism. It combines different -omics techniques such as transcriptomics and proteomics with saturated mutant collections.[1]
The suffix -ome as used in molecular biology refers to a totality of some sort; it is an example of a "neo-suffix" formed by abstraction from various Greek terms in -ωμα, a sequence that does not form an identifiable suffix in Greek.
Current usage[edit]
Main article: List of omics topics in biology
Many “omes” beyond the original “genome” have become useful and have been widely adopted by research scientists. “Proteomics” has become well-established as a term for studying proteins at a large scale. "Omes" can provide an easy shorthand to encapsulate a field; for example, an interactomics study is clearly recognisable as relating to large-scale analyses of gene-gene, protein-protein, or protein-ligand interactions. Researchers are rapidly taking up omes and omics, as shown by the explosion of the use of these terms in PubMed since the mid '90s.[10]
Genomics is the new science that deals with the discovery and noting of all the sequences in the entire genome of a particular organism. The genome can be defined as the complete set of genes inside a cell. Genomics, is, therefore, the study of the genetic make-up of organisms.
Determining the genomic sequence, however, is only the beginning of genomics. Once this is done, the genomic sequence is used to study the function of the numerous genes (functional genomics), to compare the genes in one organism with those of another (comparative genomics), or to generate the 3-D structure of one or more proteins from each protein family, thus offering clues to their function (structural genomics).
In crop agriculture, the main purpose of the application of genomics is to gain a better understanding of the whole genome of plants. Agronomically important genes may be identified and targeted to produce more nutritious and safe food while at the same time preserving the environment.
Genomics is an entry point for looking at the other ‘omics’ sciences. The information in the genes of an organism, its genotype, is largely responsible for the final physical makeup of the organism, referred to as the “phenotype”. However, the environment also has some influence on the phenotype.
DNA in the genome is only one aspect of the complex mechanism that keeps an organism running – so decoding the DNA is one step towards understanding the process. However, by itself, it does not specify everything that happens within the organism.
The basic flow of genetic information in a cell is as follows. The DNA is transcribed or copied into a form known as “RNA”. The complete set of RNA (also known as its transcriptome) is subject to some editing (cutting and pasting) to become messenger-RNA, which carries information to the ribosome, the protein factory of the cell, which then translates the message into protein.
|
Figure 1. Genes, proteins, and molecular machines |
|
|
|
Source: U.S. Department of Energy Genomes to Life Program, http://doegenomestolife.org |
The International Rice Genome Sequencing Project
|
|
This ongoing genomic research in rice is a collaborative effort of several public and private laboratories worldwide. This project aims to completely sequence the entire rice genome (12 rice chromosomes) and subsequently apply the knowledge to improve rice production.
In 2002, the draft genome sequences of two agriculturally important subspecies of rice, indica and japonica, were published. Once completed, the rice genome sequence will serve as a model system for other cereal grasses and will assist in identifying important genes in maize, wheat, oats, sorghum, and millet.
For more, visit http://rgp.dna.affrc.go.jp/IRGSP
Proteomics
Proteins are responsible for an endless number of tasks within the cell. The complete set of proteins in a cell can be referred to as its proteome and the study of protein structure and function and what every protein in the cell is doing is known as proteomics. The proteome is highly dynamic and it changes from time to time in response to different environmental stimuli. The goal of proteomics is to understand how the structure and function of proteins allow them to do what they do, what they interact with, and how they contribute to life processes.
An application of proteomics is known as protein “expression profiling” where proteins are identified at a certain time in an organism as a result of the expression to a stimulus. Proteomics can also be used to develop a protein-network map where interaction among proteins can be determined for a particular living system.
Proteomics can also be applied to map protein modification to determine the difference between a wild type and a genetically modified organism. It is also used to study protein-protein interactions involved in plant defense reactions.
|
|
For example, proteomics research at Iowa State University, USA includes:
- an examination of changes of protein in the corn proteome during low temperatures which is a major problem for young corn seedlings;
- analysis of the differences that occur in the genome expression in developing soybean stressed by high temperatures; and
- identifying the proteins expressed in response to diseases like soybean cyst nematode.
Metabolomics
Metabolomics is one of the newest ‘omics’ sciences. The metabolome refers to the complete set of low molecular weight compounds in a sample. These compounds are the substrates and by-products of enzymatic reactions and have a direct effect on the phenotype of the cell. Thus, metabolomics aims at determining a sample’s profile of these compounds at a specified time under specific environmental conditions.
Genomics and proteomics have provided extensive information regarding the genotype but convey limited information about phenotype. Low molecular weight compounds are the closest link to phenotype.
Metabolomics can be used to determine differences between the levels of thousands of molecules between a healthy and diseased plant. The technology can also be used to determine the nutritional difference between traditional and genetically modified crops, and in identifying plant defense metabolites.
|
Figure 2. Example of a metabolic network model for E. coli |
|
|
|
Source: U.S. Department of Energy Genomes to Life Program, http://doegenomestolife.org |
|
Figure 2. Example of a metabolic network model for E. coli |
|
|
|
Source: http://biotech.nature.com |
Genomics provides an overview of the complete set of genetic instructions provided by the DNA, while transcriptomics looks into gene expression patterns. Proteomics studies dynamic protein products and their interactions, while metabolomics is also an intermediate step in understanding organism’s entire metabolism.
Glossary
Chromosome: a grouping of coiled strands of DNA, containing many genes.
DNA (deoxyribonucleic acid): a molecule found in cells of organisms that encodes genetic information.
Gene: a biological unit that codes for distinct traits or characteristics.
Genome: the complete set of genes in a cell.
Genotype: the genetic constitution of an organism.
Metabolome: complete set of low molecular weight compounds in a cell at a given time.
Phenotype: the physical appearance/observable characteristics of an organism.
Proteome: complete set of proteins in a cell at a given time.
RNA (ribonucleic acid): a molecule, derived from DNA by transcription, that either carries information (messenger RNA), provides sub-cellular structure (ribosomal RNA), transports amino acids (transfer RNA), or facilitates the biochemical modification of itself or other RNA molecules.
References
- Genomics and Its impact on Medicine and Society. A 2001 primer. http://www.ornl.gov/TechResources/Human_Genome/publicat/primer2001/1.html
- Primer on Molecular Genetics. http://www.ornl.gov/TechResources/Human_Genome/publicat/primer/primer.pdf
- Plant Sciences Institute Update. Iowa State University. October 2001. Volume 2 No.1.
- Genome Projects. http://www.tigr.org/tdb
- Meet the ‘omics’ 2003 Agbiotech Infosource. Saskatchewan Agricultural Biotechnology Information Centre, A service of Ag-West Biotech Inc.
Reference
1. https://en.wikipedia.org/wiki/Genomics
2. https://en.wikipedia.org/wiki/Omics
3. http://www.isaaa.org/resources/publications/pocketk/15/default.asp
4. http://geneticscertificate.stanford.edu/courses/genomics-and-the-other-omics